Basic Volume Analysis
Nearly any anatomical image can be analyzed for CSF volumes, but those which are most trivial to analyze are heavily T2-weighted images. Heavy T2 weighting suppresses any non-CSF components of the image, and make volumetric analyses straightforward.
We use ImageJ in this basic analysis walkthrough because it is free, available on multiple platforms, and allows the user to modify the images (when needed to threshold parenchyma and CSF). Much more efficient analysis workflows are possible with supervised and unsupervised algorithms if large numbers of images need to be processed.

Download Example Images
Software Prerequisites
We recommend the following software:
-
Download ImageJ (free for MacOS and Windows) for post-processing analysis of t2-weighted images
-
Download Horos (MacOS users) to view DICOM images
Steps for Measuring Volume With ImageJ
-
Open ImageJ.
-
Open the t2 weighted image you want to analyze (select File → Open)
-
Obtain the voxel dimensions. If needed, you can obtain these from imageJ (Select Image →Properties), and record the voxel size (width, height, depth). In the example image they are 0.479 x 0.479 x 4.00 mm.
-
Open the window and leveling tool (select Image →Adjust →Window/Level).
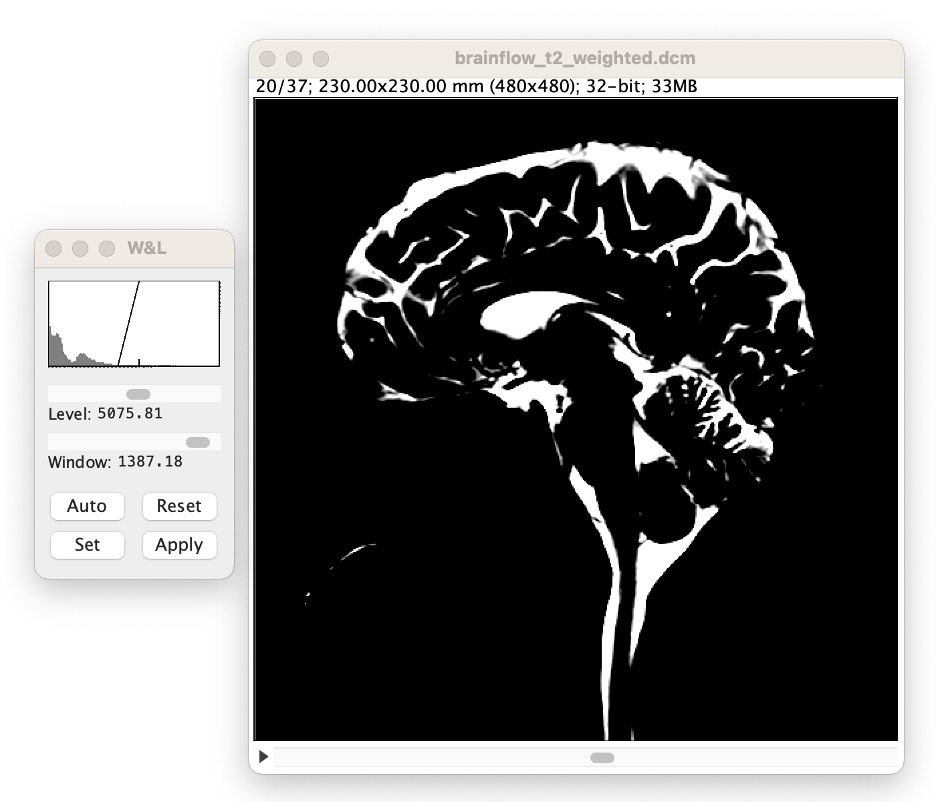
The W&L window a histogram of intensities in the current image slice (x-axis = image value, y-axis = number of voxels), and a 'ramp' or 'triangle' indicating what values are shown on the screen. The left side of the ramp is the lowest displayed value. All voxels lower than this value will be displayed as black. The right side (indicated with a tick mark) is the highest displayed value. All voxels higher than this value will be displayed as white. Note if you select "apply" the values in the image will be changed. We recommend you do not select "Apply". Thus, only the way values presented on the screen will be changed during this process.
-
Navigate through the images by using the scroll bar at the bottom of the image.
-
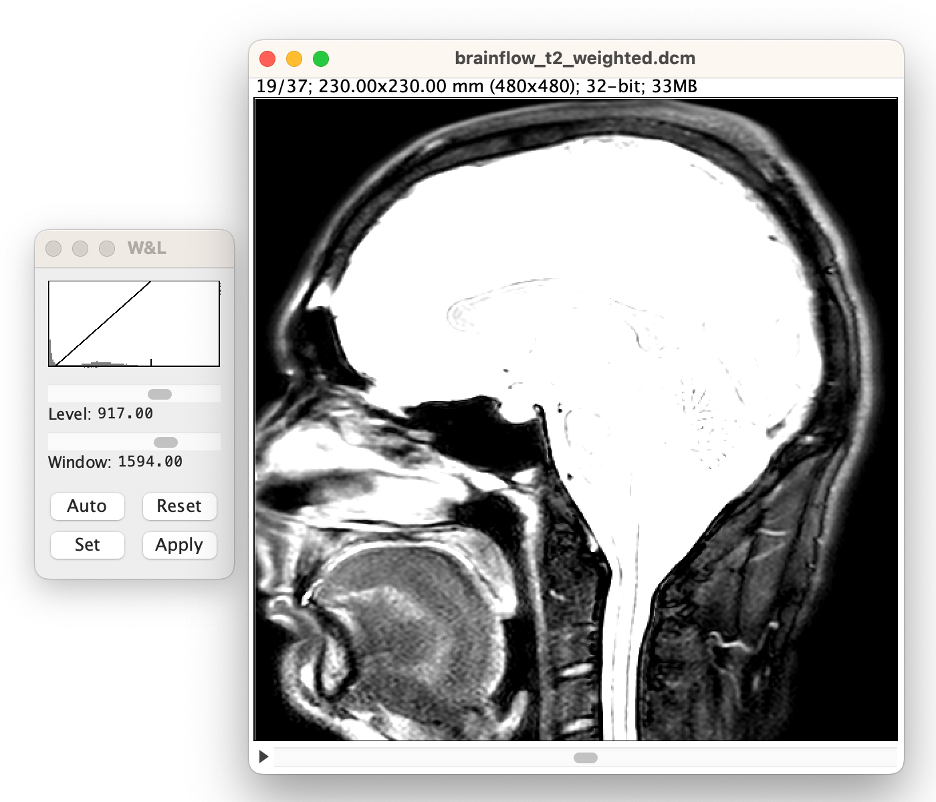
Stop at approximately the middle of the image.
-
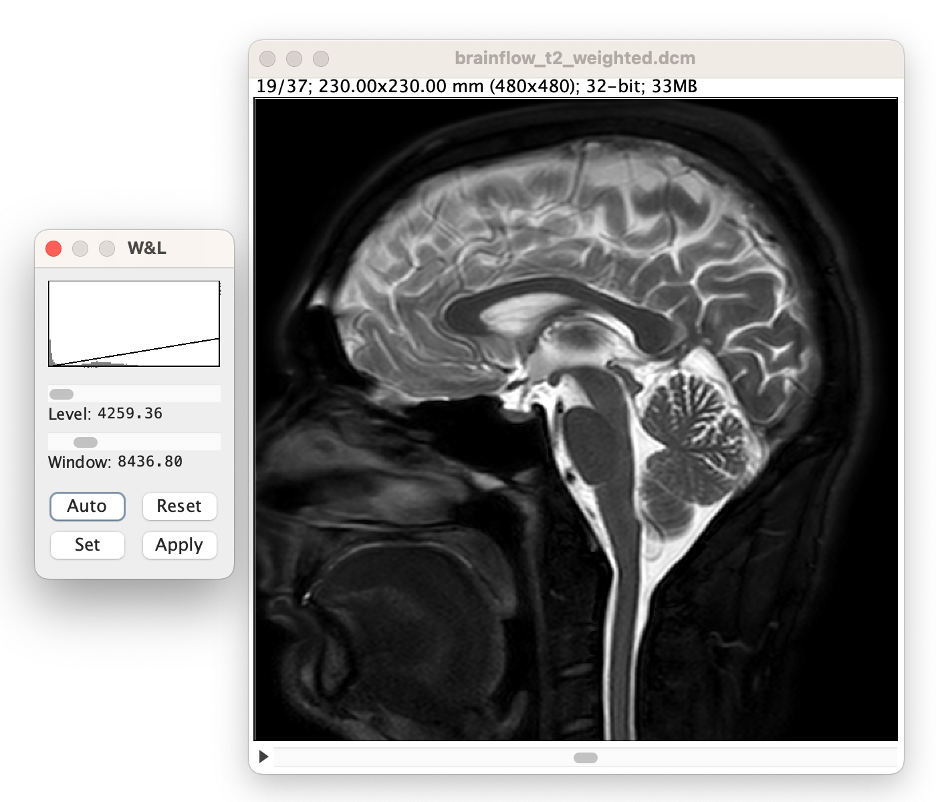
Note that most of the brain will be too bright to see. Use the window and leveling tool to visualize the brain better. (select Auto in the W&L menu)
-
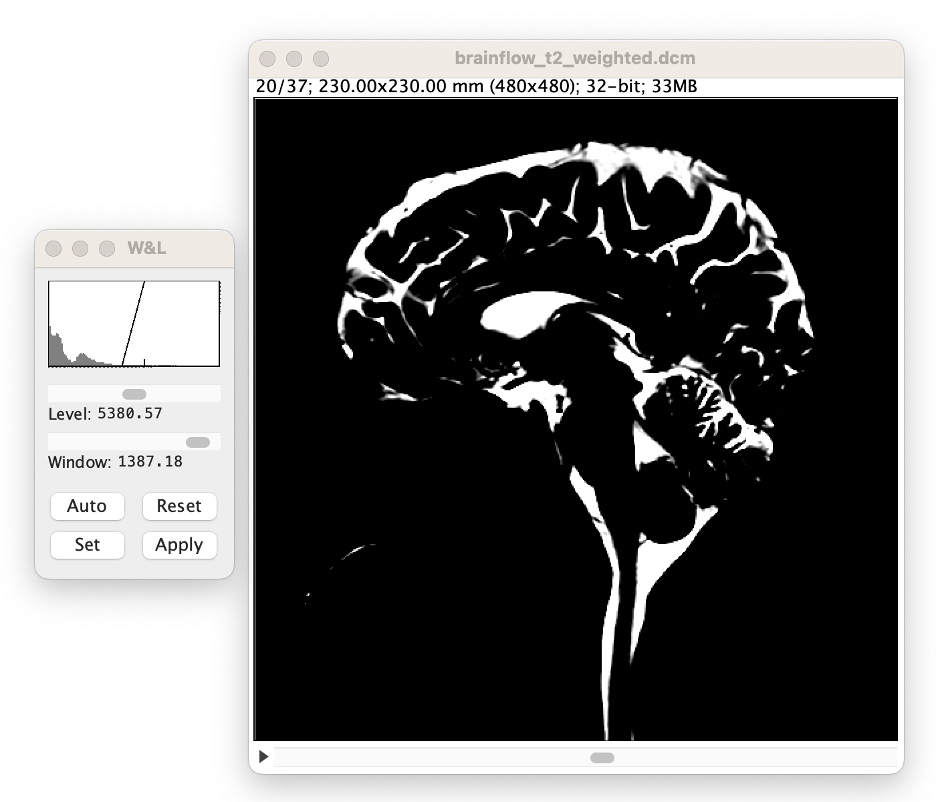
Adjust the 'window' slider until the ramp is approximately 10% the width of the histogram. Note: this approximate value does not affect CSF volume accuracy, it merely aids in the volume analysis process.
-
Adjust the 'level' slider. Note: this this affects CSF volume accuracy.
-
Moving it to the left (lower levels) will decrease the likelihood of missing CSF, but may also select some parenchyma and thus overestimate CSF volume.
-
Moving it to the right (higher levels) will increase the likelihood of missing CSF, but will deselect parenchyma and thus may underestimate CSF volume.
-
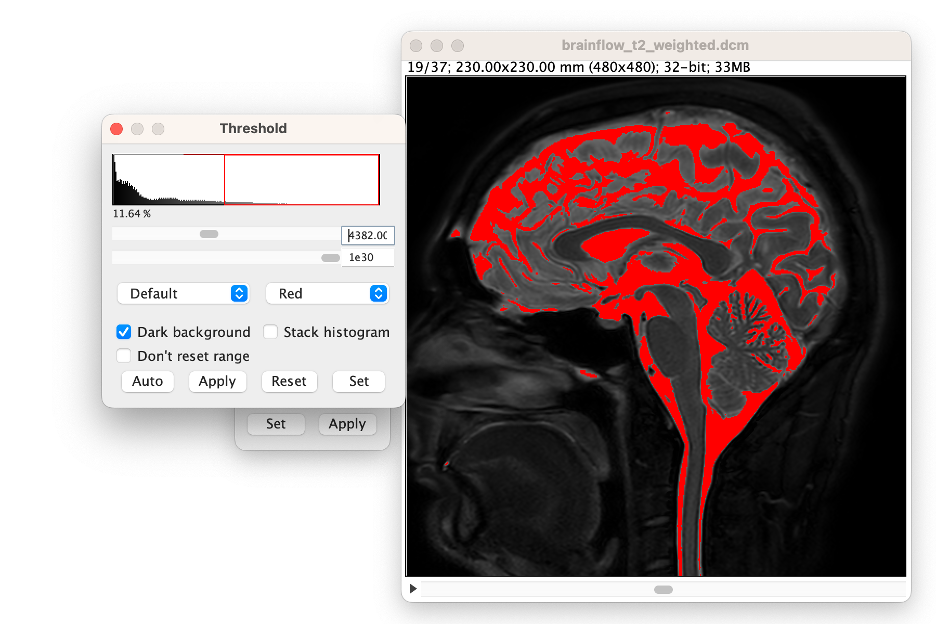
Compute the threshold value. You can now compute the value for your threshold process. We recommend using the lower point of the 'ramp' which can be easily calculated by taking the level value (displayed in W&L menu) and subtracting half the window value (displayed in W&L menu). In this example, the threshold value is 4382.
-
Refine and View the Threshold Value (select Image →Adjust →Threshold). In the Threshold window, select "Dark Background," then enter the computed value into the upper slider bar's window. Note: you can refine the value by adjusting the slider bar.
-
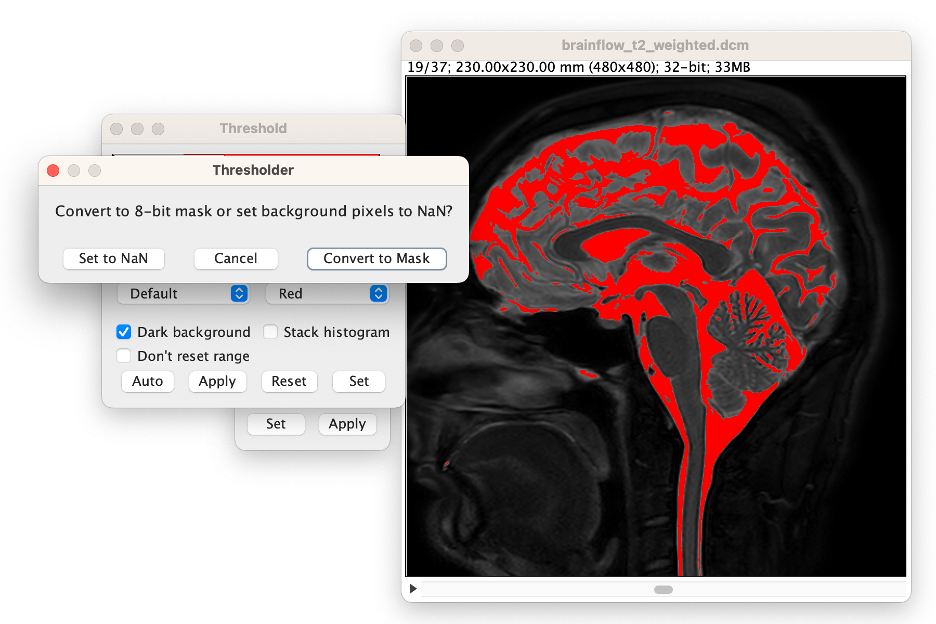
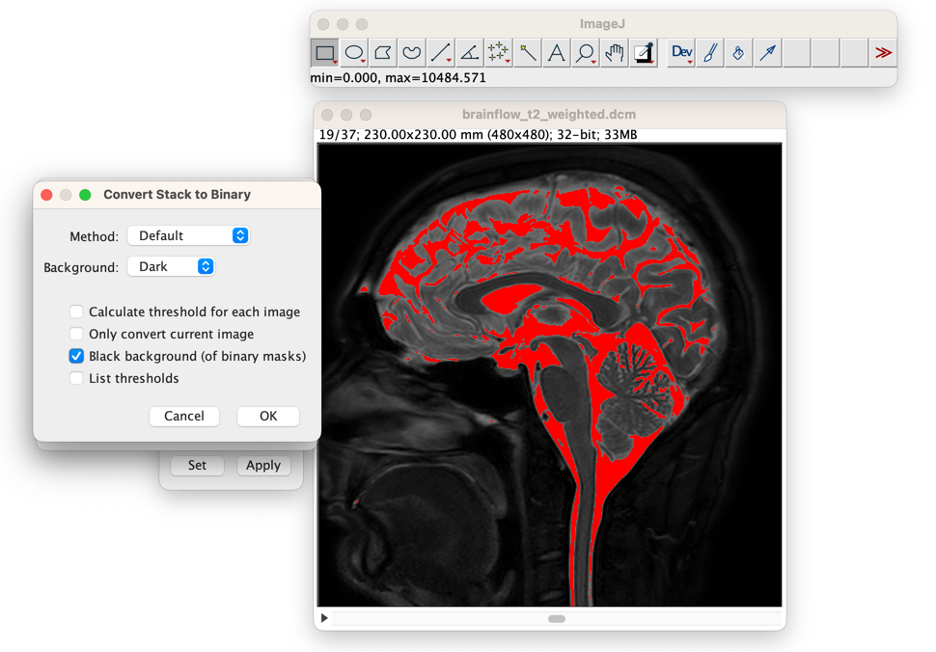
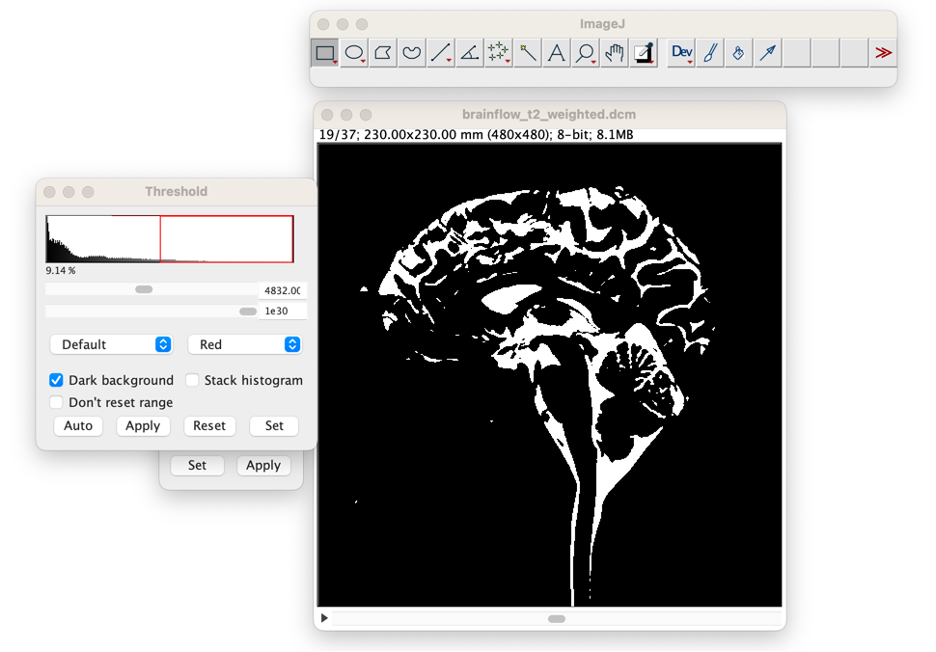
Apply the Threshold Value. In the Threshold window, select "Apply", in the first window that opens then select "Convert to Mask", and in the second window that opens select "Background: Dark" deselect "Calculate threshold for each image," and select "Black Background", then press "OK."
-
Close Threshold Window and the W&L window. The image is now an 8-bit image where the non-CSF regions have the value 0, and the CSF regions have the value 255.'
-
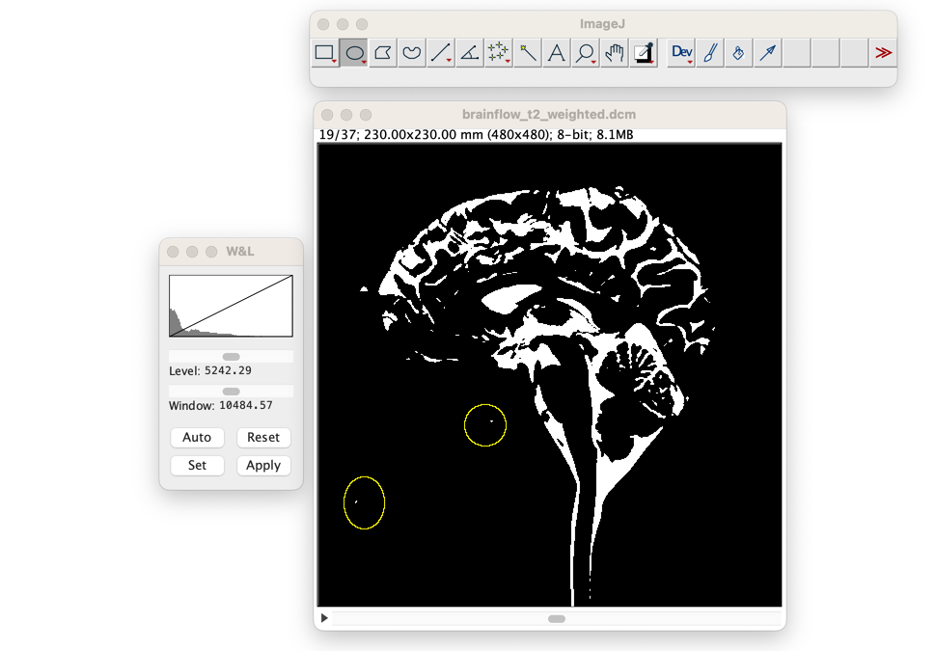
Manual Adjustments to the CSF Volume. Select one of the region of interest (ROI tools) from the ImageJ toolbar. In this example we selected the ellipse tool. Place ellipses around any regions that should not be CSF. Note: holding 'shift' enables the user to place multiple ellipses.
-
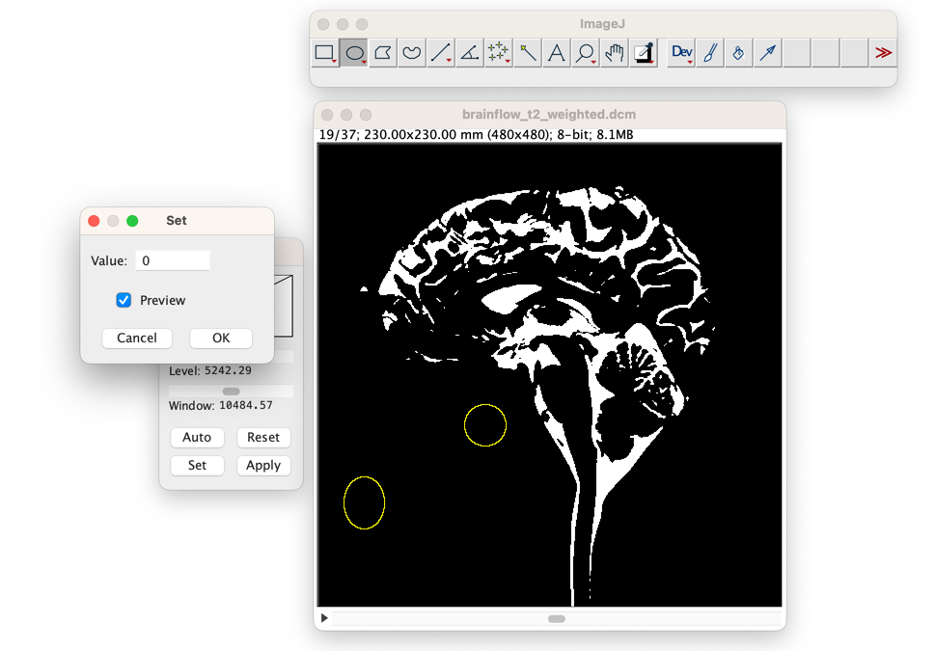
Fill regions of interest. If you wish to only remove CSF from this one slice, then you can press "delete." Otherwise if you wish to remove the ROI from all slices, or if you wish to add the Roi location back to CSF, then (select Process →Math →Set). Then change the value to "0" (for not CSF, or "255" for CSF) and select "preview" to see the change. Press "OK" when you wish to apply the adjustment. If this region should be changed in only this image, then press "No' In the subsequent window. If this region should be changed in the entire volume of imaging, then press "Yes." Repeat this process on all slices until you are satisfied with the results.
-
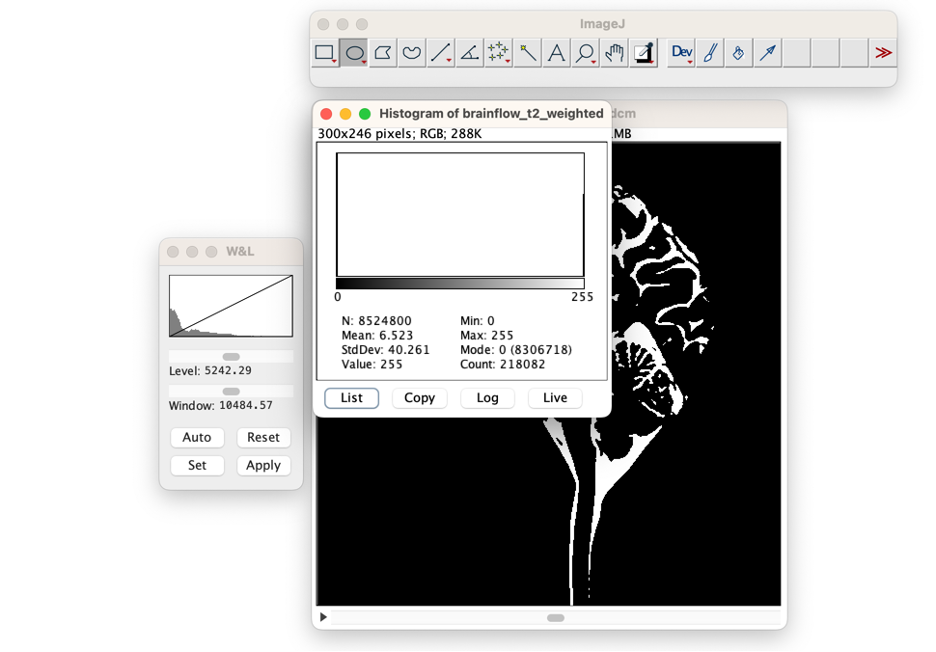
Count CSF Voxels. (select Analyze →Histogram). When a window opens asking if you want to include all images, select "Yes." Put your cursor over the bar indicating bright (CSF) voxels at "255". The number of CSF Voxels will be listed as "Count." In the example image the count is 218,082 voxels.
-
Compute CSF Volume. Using the voxel sizes (0.479 x 0.479 x 4.00 mm in example image) and number of CSF voxels (218,082 in example image) you can multiply to obtain the CSF volume: 0.479 x 0.479 x 4.00 x 218,082 = 200 milliliters. Note that 1 mm cubed is equal to 0.001 milliliters.
